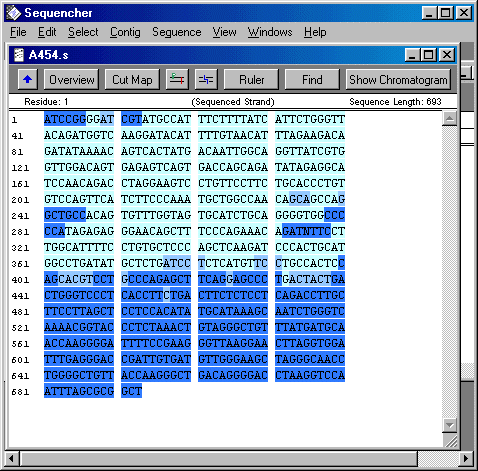
So this last week I focused on organising and cleaning up sequences in a Sequencher program. I had to find SNPs and label them by hand. Since I was reviewing 20-ish different 200 to 1000 bp sequences for 2 to 24 individuals each. THat's a lot of letters. So I didn't quite finish, but I hope to finish up on Monday. Not only did I label SNPs, I also cut off the ends of each sequence that were not reliable. Sequencher actually reads the chromatogram and translates the flourescent waves into four letters, the letters that encompass the genetic code (C, A, T, and G). Sometimes, though, Sequencher makes a bad call. When it miscalls a base or is not sure about a base, I have to fix that by changing the sequence. Once I have cleaned up all the sequences so that all sequences of the same primer pairs are the exact same length, then I will get help from some other people in the lab on analyzing the data. The picture is what the sequence looks like on sequencher. Dark blue bases mean that Sequencher is less sure about the base call, whereas light blue bases mean that Sequencher is pretty certain that the base is as it calls it.
No comments:
Post a Comment