Wednesday, August 1, 2007
Belated (but oh so necessary) Goodbye
Tuesday, July 31, 2007
Final wrap up
Thanks so much to all of those who kept posting to the blog throughout the summer. I think it was an overall success, and we'll be adding it as a requirement to next year's program.
Once again, great job everyone! Please keep us updated about your career goals and where you end up for college! Good luck this next year and enjoy the rest of the summer!
Thursday, July 26, 2007
Going out with a bang!
Today was transplant day for one of the two patients waiting right now--very exciting!!
I was able to observe in the operating room with Dr. Markert, Julie (who works in the lab) and two other students (Dave and Ashley). After we carried over all the slices of thymus in a plastic bin and suited up in bunny-suits, hair and shoe covers, and masks we entered the operating room. A resident was actually performing the surgery with one or two doctors and several nurses, and us. It was a pretty full house but luckily we were in a pretty big room. They had started to put the central line into the baby when we arrived and it took them a while to get it positioned correctly. They start by putting a needle near the neck and then using it to guide a wire through a vein all the way to the heart, and then thread a tube over the wire and pull the wire out. They used x-rays to watch the position on the wire which we could watch on a screen. The central line will be used to give the patient calcium and nutrients while he's waiting for stomach surgery. They then started the transplant procedure by making an incision on both thigh muscles. Fortunately, one of the students was able to recognize at this point that they were not meant to be a surgeon and had to go sit down right away. I went out into the hall when the person fainted. Luckily, we were already in a hospital and some nice doctor brought us a wheelchair and some food. And so...i missed most of the actual transplant surgery. I was able to go back in the operating room just in time to see the last thymus slice inserted and watched them sew it up. Needless to say, it was a very interesting day at the hospital all around and a high light of the summer!
I'm so grateful for this opportunity, i can't imagine a better way to have spent time working this summer. Everyone in the lab made it such an enjoyable and amazing experience. Thanks to everyone in the group too, it's been fun!
Wednesday, July 25, 2007
the end, oh sorrow
~love from linda
Monday, July 23, 2007
The End
Results
Thursday, July 19, 2007
Formatting
Sunday, July 15, 2007
Killifish Eggs and Hatchlings
This has been harder then we first expected. The first contaminant we exposed them to was Ammonium Chloride. We tried three dose responses with killifish eggs. In the first experiment, we tried doses ranging from 7.5 mg/L to 150 mg/L. The fish had no reaction and continued with development as usual. We tried another dose ranging from 250 mg/L to 1500 mg/L and once again the killifish eggs showed no response. We did a final, more extreme experiment with doses ranging from 2,500 mg/L to 10,000 mg/L. When the eggs showed no response to this as well, we came to the conclusion that the killifish are protected inside their eggs and that the ammonia does not penetrate the egg. We later did experiments in killifish hatchlings which did show a response to the ammonia. I never thought I'd be happy to say that something died, but after watching many fish eggs survive the ammonia doses, I was glad to deliver the news that hatchlings did show the response we expected to the ammonia and that we would be able to continue on with our research.
Although, this presented an obstacle in continuing with my research of whether Elizabeth River killifish have a fitness cost associated with their adaptation to PAH's. We could no longer do research in eggs because the pesticides we were exposing them to proved to be similar to the ammonia in the sense that the killifish eggs showed no response. Instead we would need to do our research with hatchlings. Killifish eggs take two weeks to hatch. A large quantity of them, 350, will be ready from hatching tomorrow but this set us back a bit in data collection. We will conduct the majority of our research on these hatchlings next week using the data that we have gathered in the last few weeks with the dose response experiments, to know what the correct doses are to reach the LC 50. It will be another busy week, ending of course with the release of the final Harry Potter book. I know many of us in the program are looking foreward to that.
BuSy Week With the ADHD samples!!!!
Sequencher
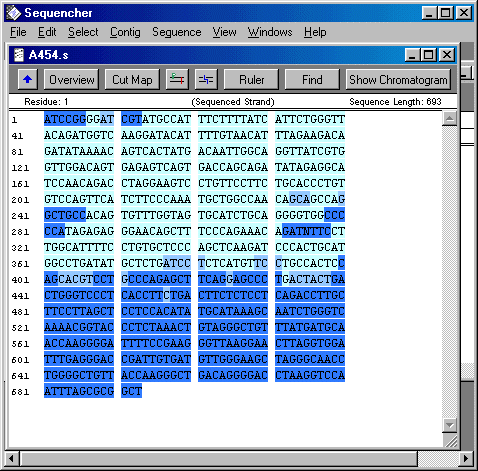
So this last week I focused on organising and cleaning up sequences in a Sequencher program. I had to find SNPs and label them by hand. Since I was reviewing 20-ish different 200 to 1000 bp sequences for 2 to 24 individuals each. THat's a lot of letters. So I didn't quite finish, but I hope to finish up on Monday. Not only did I label SNPs, I also cut off the ends of each sequence that were not reliable. Sequencher actually reads the chromatogram and translates the flourescent waves into four letters, the letters that encompass the genetic code (C, A, T, and G). Sometimes, though, Sequencher makes a bad call. When it miscalls a base or is not sure about a base, I have to fix that by changing the sequence. Once I have cleaned up all the sequences so that all sequences of the same primer pairs are the exact same length, then I will get help from some other people in the lab on analyzing the data. The picture is what the sequence looks like on sequencher. Dark blue bases mean that Sequencher is less sure about the base call, whereas light blue bases mean that Sequencher is pretty certain that the base is as it calls it.
For the last two weeks, I have been continuing to work on my Toluene experiments. Turns out that the results were not as promising as we had hoped since the E. coli conjugates do not seem to be breaking down the Toluene. :-( I have been testing different conditions to see if the toluene will degrade in more stressful environments, and some seem to be working a little better than the others. One possibility is that the original stock solution has been contaminated, since it has been sitting in the refridgerator for a while now. We should probably make a new stock solution on Monday. Hopefully Ruoting and I will be able to find advantageous conditions for the E. coli soon. :-D
Annie
Harry Potter and the Deathly Hallows
5 days. 4 hours. 27 minutes. 07 seconds.
Tuesday, July 10, 2007
The only slight problem this poses is that I will now have a more difficult time in trying to choose which project to focus on for my poster presentation. Additionally, I am worried that I will be short on time and will have a harder time acquiring all the data when my time is split three-ways. In eight minutes now, I'll be rock-climbing with some people from our lab for the first time ever. I hope to survive so I can at least finish the last book of the Harry Potter series. I'll show you my scrapes and cracked jaw bone tomorrow. Chow!
Monday, July 9, 2007
Making a Slide
 So. Here's how I make a slide.
So. Here's how I make a slide.1. Line up embryos with a probe (pointy thing).
2. Put them on a slide (see the little white smudge in the middle?).
We put them in halocarbon oil so they can still breathe!
3. Put the slide in the microscope.
4. Turn on the laser. (488nm = blue)
5. Viola! (10x lens)
This is basically what I do for my experiment. Today, I did a time lapse series. I took pictures every hour to see how GFP expression varies over time. Hopefully, I'll be able to fit a line or a curve to my data points tomorrow.
Drosophila Drosophila Drosophila (try saying it 3 times fast)
Because my project compares different pictures taken at different times, we needed to figure out a way to make a "control" in each picture. The pictures are different because the embryos fluoresce under a 488nm laser. That laser can vary in power and make 2 different fluorescing embryos look the same. Different embryos also require different laser powers to create 'optimal' pictures--not too bright or too dim.
The first thing we tried was fluorescent beads. The idea made sense: squirt some of these 6um beads (embryos are about 150 micrometers by 400um) onto each slide. Take pictures. Use the ratios of bead brightness from each picture to compare embryo brightnesses (ex: in one picture, the beads have a pixel value of 1. in the other, 2. to compare, double the brightness of the embryo in picture one).
But in practice, this was not so easy. First of all, the beads needed to be kept in the dark and at around 4C all the time. This meant I had to do all the squirting in the dark in the 'cold room' where my lab works with proteins. Needless to say, it was pretty hard. I would either miss the spot on the coverslip (if the embryos and beads weren't in the same frame for pictures, it wouldn't work), the oil I was injecting into would freeze (you put embryos in halocarbon oil to keep them alive under a coverslip), or if I did get the beads right, they would mess themselves up. Once, the computer measured the 3% beads as WAY brighter than the 30%. So beads weren't so helpful. (one of the grad students had ordered them a long time ago to do what I am now, but got frustrated and decided it wasn't worth the time)
But luckily we found a paper. And it described a method that did not include beads. Our new method allows us to adjust laser power for each embryo and then normalize the values to make them comparable. Basically, you view the results as if you had taken each picture at 1% laser power. First, you raise the laser power to make the embryo you are viewing a certain brightness. You take the picture, take the mean brightness of the embryo (with a histogram on the computer) and divide by the laser power you used (ex: mean brightness=100 laser power=25% normalized brightness=4).
I'm really enjoying the new method we found for two reasons: 1) I get to use the really fancy 510 confocal microscope (I will post pictures) 2) I feel like I got to have creative/intellectual input when it came to how we were going to implement it. No one in the lab has done it before, so we kind of got to start from scratch.
Anyway, now I'm going to put up some pictures from a few weeks ago. Next time: my side projects-- inverse PCR and gene mapping!
FAmily Visit
I was able to observe my first family visits on a Friday. So far, that was probably one of the highlights of my experience with the program. I watched as the entire family of four completed a series of behavioral and psychological tests as well as developmental histories to see if they could be diagnosed with ADHD. There are three rooms in the clinic area: a computer room, an interview room, and a blood room. One member of the family goes into the computer and interview room at a time. In the computer room, letters appear on the computer simultaneously at varying time intervals and every time a letter, other than “X”, appears, the participant has to push the space bar. This test takes place for exactly fourteen minutes. After a while, one of the young participants that I watched became bored of the test, so he continuously and randomly just began pushing the space bar, increasing his impulsitivity score. The interview room is where the psychologist assesses them and gives them an IQ test by asking several questions as well as by allowing them to complete spatial and hands-on tasks. Finally, after everyone in the family completes these tests, there blood is drawn and set to the DNA bank. Unfortunately, one of the children refused to get a blood draw. Overall, the entire family visit lasted for a period of three hours and thirty minutes.
 them on a pretty regular basis. This week there should be enough cells to harvest them and extract RNA. I also observed the extracting of PBMC's (peripheral blood mononuclear cells) which contain the lymphocytes and monocytes (cells of the immune system). When spun at high speeds the blood separates into different levels according to density. The lymphocytes and monocytes form a buffy coat under the layer of plasma that you pipette out. It's important to get good separation of the blood for clear results in many of the tests in the lab. Once the pbmc's were separated Jie showed me how they select the T cells (a type of lymphocyte) from the sample magnetically. T cells express CD3 on the cell surface. The white blood cells are mixed with CD3 antibodies that are attac
them on a pretty regular basis. This week there should be enough cells to harvest them and extract RNA. I also observed the extracting of PBMC's (peripheral blood mononuclear cells) which contain the lymphocytes and monocytes (cells of the immune system). When spun at high speeds the blood separates into different levels according to density. The lymphocytes and monocytes form a buffy coat under the layer of plasma that you pipette out. It's important to get good separation of the blood for clear results in many of the tests in the lab. Once the pbmc's were separated Jie showed me how they select the T cells (a type of lymphocyte) from the sample magnetically. T cells express CD3 on the cell surface. The white blood cells are mixed with CD3 antibodies that are attac hed to an iron molecule. These micro beads only attach to the cells expressing CD3 and the whole sample is run through a filter with a magnet. It was cool to see the process from beginning to end starting with whole blood and ending with a specific type of lymphocyte.
hed to an iron molecule. These micro beads only attach to the cells expressing CD3 and the whole sample is run through a filter with a magnet. It was cool to see the process from beginning to end starting with whole blood and ending with a specific type of lymphocyte. The DLC is always fun, and this week will be a little different. We are "shaping" two new female mongoose lemurs (Guadalupe and Eduardo)-teaching them how to use the touch screen (contrary to what many believe, just because Rosalita may have watched Eduardo do the experiment, she still has no idea how to even approach the computer). Aristides will also start again with the Risk paradigm after being out sick for quite a while. Aristides will be so excited, as whenever we walk by with the machine he thinks it's his turn and is ready to go! Finally he'll be able to. Also thanks to Aristides (a ringtail) I have data to use for my project. Aristides has finished the experiment that Teres is now running (that I am using for my project).
Hope everyone had a great 4th and a weekend! This summer is going by just way too fast....
ADHD
Tuesday, I worked on a Taqman plate and two PCR plates. After running them on the machine, I had to wait until they finished, so that they could be tested on an agarose gel. After lunch, I tested the samples on the gel. My results were better than my results on the previous day. So, I completed two more PCR plates using a different primer, and ran them on the PCR machine. Then Chris taught me a new method using Sephadex to filter the DNA from our PCR. It was fun being able to learn a new method. This was a gel like substance that allows the DNA to separate from the excess materials used during the PCR reaction. After that, I worked on my blog.
I had a wonderful Independence Day celebration with my friends on Wednesday and returned to work on Thursday. I felt so excited to receive my first paycheck. When I came to the lab, I made an agarose gel and ran my previous PCR samples on it. It actually worked out well. Afterwards, I made another PCR plate prior to lunch. I made another PCR plate after lunch and I also began labeling my ADHD plates. I then waited for my PCR plates to finish running so that they could be tested on an agarose gel.
Friday was a great day! I began my day by making a PCR plate. I then ran my PCR agarose gel that I made earlier in the week. Fortunately, all my samples worked. Next, I ate lunch early because I was invited to observe a family visit. This is when the proband as well as the entire family come into the clinic area to be tested for ADHD.
Sunday, July 8, 2007
Sequences available!
Saturday, July 7, 2007
Mimbulus Mimbletonia
 So the peaks portray how many base pairs long different alleles are. However, it's not as easy as it sounds because there are deceitful little peaks that try to trick you into believing that they're real - but really, they're just imposters... Anyway, I've been trying to tackle this program, but it's been putting up a good fight. But I'm determined to conquer it…somehow…
So the peaks portray how many base pairs long different alleles are. However, it's not as easy as it sounds because there are deceitful little peaks that try to trick you into believing that they're real - but really, they're just imposters... Anyway, I've been trying to tackle this program, but it's been putting up a good fight. But I'm determined to conquer it…somehow…
In the greenhouse, I'm still monitoring the germination and flowering of the M. tilingii and M. guttatus; however, many of the tilingii were looking slightly puny, so we ended up replanting some of them.
But thankfully, some of the paired guttatus individuals started flowering, and I began taking down morphological data. We decided on comparing and contrasting flowering time, basal width, overall height, number of vegetative stems, stem thickness between different nodes, internode lengths, leaf length/width, corolla length/width, stigma length, anther length, calyx length/width, and number of flowers after a certain time period.
We're also planning on measuring stomatal density and perhaps (hopefully, if all goes well) trichome density/morphology. Measuring these two traits is really interesting! So for stomatal density, you take casts of the underside of leaves using dental impression material and stick it on a microscope slide. When that dries, you spread some clear nail polish on the casts and make leaf impressions from the nail polish peels. You put those peels on another microscope slide and examine it under a microscope, and you can see individual plant cells, including the guard cells that control stomatal opening/closing! Super cool! Also, I've been wanting to examine more closely the tiny little hair-like structures that are found on both the tilingii and guttatus. Here's an amazing picture of a tilingii individual in which the trichomes are really easy to see (taken by Carrie):
 Amazingly, some of the tilingii populations (like this one) have a sticky substance on their trichomes. It's hypothesized that this glandular exudate is a mechanism for resistance to insect herbivory, insulation, etc. Anyway, we looked at the trichomes on both the tilingii and guttatus and discovered that for the individuals that we examined, the trichome morphology was different between the two species
Amazingly, some of the tilingii populations (like this one) have a sticky substance on their trichomes. It's hypothesized that this glandular exudate is a mechanism for resistance to insect herbivory, insulation, etc. Anyway, we looked at the trichomes on both the tilingii and guttatus and discovered that for the individuals that we examined, the trichome morphology was different between the two species
Oh, and I almost forgot! I was reading Harry Potter and the Order of the Phoenix, and there's this part that describes a plant that Neville has called "Mimbulus mimbletonia". Doesn't "mimbulus" sound eerily similar to "mimulus"? Anyway, I looked it up on Wikipedia:
"Mimbulus mimbletonia is described as "appear[ing] to be a small grey cactus in a pot, except that it was covered with what looked like boils rather than spines" (OotP 186). The genus name mimbulus may be related to the real genus mimulus, especially since those plants are used as folk remedies for "shyness, anxiety, and forgetfulness" and those are traits of Neville Longbottom."Mimulus are definitely not small grey cacti, but I thought that was a cool coincidence. Now whenever I go to the greenhouse, I feel like I'm in Professor Sprout's class! =]
Thursday, July 5, 2007
Romeo oh Romeo....

A post will follow later in the week about my lab week, but I have a few new photos I wanted to put up. Evan showed us this adorable situation. Here are a few pictures of Romeo (diademed sifaka) and the squirrel, a photo of Chloris' baby (ringtail) and one of Eduardo(mongoose). Some of them are a bit fuzzy because I cant use flash.
Monday, July 2, 2007
lab raiding
I'm also looking at the subclasses of a particular antibody that patients with HITT have in their plasma. This week I finished optimizing the ELISA for that part so I get to begin collecting data from more samples. That was very exciting because it will *hopefully* get in Dr. Arepally's new paper if the results turn out well.
This week, our lab killed some of the mice they do work with. I didn't go to see because I don't have clearance - I probably wouldnt have gone anyways, it sounds so gruesome. We also raided another lab that was closing so I spent a lot of time carrying stuff back and forth. At one point we have TEN CHAIRS in our tiny lab along with the piles of other stuff we found. It was fun finding all these cool gadgets and chemicals that we needed too.
Babysitting Jurkat Cells...

I also started doing more work in the lab this week. My job was to recover a human T cell line called Jurkat cells. The Jurkat cells needed a specific medium which I put together. Then came "waking them up" which involved finding the right tube in the liquid nitrogen, and heating them quickly in a water bath. The cells are very touchy so you have to be careful and move quickly. I counted the alive cells using a hemocytometer. Every day I checked the cells under a microscope to make sure they were happy and growing. There were a lot of dead cells floating around in the flask but hopefully they don't affect the living ones.
The experiment is working! :-D
I really liked the AIDS talk that was given last week, and I was shocked that people with salaries over $12,000 couldn't get financial aid for therapies and medicines in North Carolina. Now I have this urge to write a letter to our congressmen about this issue... It'll probably not do any good, but I'll write it anyway. And then we can all sign it! :-)
Sunday, July 1, 2007
The little urchins are growing!

This week was a pretty interesting one. I'm knee-deep in my project, and the rest of the lab is working on obtaining DNA and other data from millions of baby sea urchins. On Monday I did a HUGE gel for 16 primersets and 12 individuals- that means I loaded 192 wells plus 8 ladders- all within 15 minutes. Beyond that time, the gel begins to get fuzzy and loaded DNA in one well might mix with another well. For this particular gel, I needed some high-tech equipment to fill the wells before the timer ran out. I got to use the collest pipette ever. It was just like a normal automatic pipette except that it had 8 tips instead of 1. This allowed me to load my gel much more quickly and efficiently. it's really fun to use, but hard also. Sometimes the pippette won't suck up or dispense the correct amount of liquid, which can be frustrating. So on Tuesday I got to go to the embryo sea urchin freezer where people were viewing the babies to see if they were developing on schedule. Unfortunately, some of the embryo cultures were infected with bacteria, which had to be eliminated without eliminating the babies. The first thought was to dump the sea water that the eggs were in since the eggs were on the bottom and the water was on top. But some of the eggs had already hatched and the babies were swimming around in the water. Then we used a filter to avoid the eggs and babies, but suction out the bacteria. When I got back to the lab, I looked at the big gel I had run the day before. Turns out one whole plate was ineffective, so I redid that on Wednesday. Thursday I did another gel, and Friday I got to work with the urchins again. By this stage, the urchins look like little floating, transparent pyramids that can swim. They're really cute, but sadly, they had to die for the sake of science... So I spent Friday morning centrifuging the eggs to the bottom of the tube, sucking out the sea water, and replacing it with buffer that instantly kills the larvae and preserves the DNA which is then used to understand more about the offspring. As the little babies grow, they will develop sac-like structures on their sides, from which will spring the future adult sea urchin. The interesting thing about sea urchins is that they are born twice- once from an egg, and then again from these sac-like structures. Why? Nobody knows...
Would risk be different in a female dominated society?
Something interesting we talked about at lab meeting, which I then continued talking about with Sara and Evan:
Our lab is running risk experiments with both adults, kids and lemurs. The human adults and kids run a gain condition (they risk winning one or three versus two every time) and a loss condition (they lose one every time or risk losing none or two...). The lemurs run only the gain condition. In adults, and I believe in children, boys tend to be more risky. What I talked about with Sara and Evan was the lemurs. Because we don't run any female lemurs, we don't know if this trend would continue. It would be fascinating to see, as female lemurs are dominant (I believe that's what Evan said...). The male ringtails (eulemur catta) tend to run very safe (Aracus often runs 100% safe-two sugar pellets every time). I would love to find out what would happen if we tested female ringtails, however right now none are available.
More of Gas Chambers
This week I continued helping Eileen with her gas chambers. We started our week off by finding the flaws in the chambers. The biggest problem was that the chambers were to hot in the inside. The first time we used them in the field, the plants started to transpire and the inside of the chambers steamed up. So, we spent this week figuring out a more effective cooling method. We spiraled copper tubing around the inside of the chamber and pumped ice cold water through it. Although the water condensed on the tubing and provided the plants with some water, it was the best cooling method we could find. I also got to use the big complicated machinery to drill holes and make the chamber edges sharper once again. Next week we will take the chambers into the field once again, and hopefully we will get the results we want.
Taqman and PCR
Tuesday was a fun day also. I enjoyed the Primate Center tour. I have never seen lemurs in the past and it was amazing to see how closely they resembled humans. In the lab, Chris and I went to use the Biomek robot for transferring DNA into the 384 deep-well plates. At the same time, Robin came to my lab for a lab visit. We talked about various aspects of my project as well as the poster. After lunch, I had a meeting with Dr. Allison and Melanie. We looked at different data and statistics regarding smoking, genotypes, and polymorphisms in relation with ADHD. Amazingly, we found many significance in the data. We also discussed the status of the materials for my project. I then headed back to the lab and did two PCR plates. We actually had a small going away party for two members of the lab who are going to be relocating to Miami. We had some cake and everyone in my lab joined in. Overall, Tuesday went well.
On Wednesday, I was able to make an agarose gel. It was somewhat nerve wrecking to handle even just a small amount of ethidium bromide since it could possibly affect my future children’s DNA. It was, however, interesting how we made a gel with multiple rows of wells rather than just one. The purpose of the gel was to see if our PCRs worked. After running the gel, we took a picture of it and measured the band sizes. Fortunately my first PCRs were great. After lunch, I completed one more PCR plate, and then I proceeded to do two Taqman plates for different SNPs. I then started working on my blogs for thirty minutes. Afterwards, Chris and Dr. Allsion helped me out with my introduction.
On Thursday, I met with Dr. Allison in the morning. She informed me about the upcoming family visit for the ADHD research study patients, wherein they go through a series of tests and get their blood drawn. I am very thrilled to see that. After that, I went to my lab and finished two PCR plates. When lunch was over, I ran the samples from the plates onto the agarose gel to see if my PCR was successful. For the remainder of the day, I completed some more Taqman for patients enrolled in the hostility study.
Finally on Friday, I started planning out the methods section of my poster. Also, I searched about the various SNPs in the genes that I will be studying. I had to find its role and its functions. I made three PCR plates using different primers and ran them on the PCR machine. After that, I made an agarose gel to test my plates on. When lunch was over, I tested my samples from my PCR plates. As it turned out, my PCR turned out well. For the rest of the day, I worked on some more PCR plates and headed to the Schiciano Auditorium for the seminar speaker !! This week was a busy week, but it turned out great!
Friday, June 29, 2007
How to edit your posts
1. Log in.
2. At the bottom of the published post you want to edit, next to the 'comments' link, you'll see a pencil icon. Click on that 'Edit Post' icon.
3. That should take you back to the input screen. Click 'Publish Post' to save your changes.
Linda again
This week, I began doing some preliminary data-collection relating to my project. The mission? To gather 0 weeks data for colony diameter in AF317, MG1655, and Fufu bacteria from 5 different mouse cecums, as diameter directly correlates with rate of growth. I must say that when I started, I thought it would be simple enough: just smear some bacteria onto a plate and twiddle your thumbs for an hour or two until they finished growing. Not so. Add in several hours of prep plus thawing, vortexing, incubating, labelling, massing, extracting Fufu from cecum sacs with a two-inch pipette tip, sterilizing, autoclaving, remassing, calculating and adjusting concentrations, more vortexing, diluting, spreading, and clean-up. The most amazingly tedious aspect was the diluting, as 8 dilutions were necessary for each of the 7 bacteria and each time a new micro pipette tip was needed as well as vortexing. By the end of the diluting process, around 3 hours, I think I became a little nonchalant in the dilutions...which I think led to the fact that there grew a mysterious new strain of bacteria in one of bacteria plates. Thankfully, the rest seemed entirely normal and proliferating and usable, but from now on, I resolve to keep my wits about me and take a quick break if I start to get feckless instead of risking sabotaging all my hard work. This reminds me of a hilarious yet slightly embarrassing little episode that happened that day. It was around noon and I was busy pipetting my way along, when my PI Bill comes up and asks me how long I'll be. I tell him I still have a few left to do, so maybe 30 minutes or so. Then he says "why don't you take a break from working on that?", and then proceeds to talk to the others congregated in our little lab hall. So I plop off my chair and go to lunch. An hour later, Mary Lou, my mentor, goes "There you are! We've been looking for you!" Apparently Bill had wanted me to take a break from pipetting so he could show me how to scan and photoshop plates. Everyone had stood around hypothesizing that I had an overactive bladder and was hanging around in the bathroom, until they realized that I could not possibly be in it for that long of a time. Now whenever someone speaks to me, I make sure to paraphrase exactly what they say back...in case of a misunderstanding.
Monday, June 25, 2007
We'll be visiting your labs!
Sunday, June 24, 2007
Second Week
We disecting fish on Wednesday and Thursday morning as well. Thursday I got to see what the fish looked like once they'd cut it open to look at the liver, heart and other organs. It was fascinating, even after the fish had been dead for a few minutes, you could still see heart pumping.
Thursday afternoon and Friday I did more typical work in the lab. We looked at fish embryos under the microscope and judged their heart development after they had been induced with different chemicals. I also spawned fish again and screened the eggs, pulling out the good ones to use in my experiment. Friday afternoon I dosed those eggs, and tomorrow, Monday, I'll get to look at their development.
Second Week of ADHD Research
Tuesday was another fun day in the lab. After speaking with Dr. Allison in the morning, I went to the third floor lab of the Center for Human Genetics Building. Chris asked me to pipette the Taqman recipe unto the micro-well plates containing DNA of patients enrolled in the Hostility study. However, before doing that I first had to make the recipe by adding water, master mix, and 20x assay. It was thrilling to do nearly the entire process by myself. After having to pipette two plates, I placed them in the centrifuge and took them upstairs to the PCR machine so that they can be ran for forty cycles. I was actually able to work the machine by myself (with instructions and Chris’ assistance of course =)). Then, I had to do the Taqman again, except this time, I used a different assay. This is because I was trying to find a different single nucleotide polymorphism (SNP) in the DNA. Upon returning from lunch, I created folders on the computer for my data to be stored in when we scan the plates. I also tried to find the gene and chromosome of the SNP that I was trying to observe. When I finished, it was time to take the plates out of the PCR machine so that they can be scanned using the 7900 machine. We then went to the computer to look at the data that we produced. None of the NTC controls were amplified, so we were able to submit the data to the clinical people for analysis. Finally, to end my day, we used the Biomek robot again. Overall, I was thrilled to be able to learn new techniques.
On Wednesday, June 20, I came to my lab and proceeded to the Post PCR refrigerator to get the plates that I prepared the day before. Chris allowed me to scan the plates by myself. Fortunately, I remembered most of the process that he showed me the prior. After scanning, he asked me to do the Taqman for the C-plates for Hostility. Since not all of the micro-wells contained DNA samples, I had to look at the chart to see where to pipette the Taqman recipe. I carefully completed that tedious process and centrifuged my resulting plates. Next, I ran the plates onto the PCR machine. When I returned after lunch, I did some more Taqman on DNA from patients enrolled in the Hostility study. Upon completion, I ended up with about fourteen plates. I went upstairs to store the plates in the cold room. This was a great day of practice and precision.
I had a rewarding day at the lab on Thursday. All of the plates that I ran on the PCR machine the prior were successful. I then looked at the results on the computer and none of the NTC controls amplified. After which, Chris showed me the DNA sequencing machine, wherein he uses a restriction enzyme to find a single nucleotide polymorphism in an insertion deletion. This is completed using micro satellites. He then asked me to do some more Taqman. I finished the list of assays needed to be run. Upon completion, I felt proud that I completed about twenty micro-well plates. Moreover, I am happy to say that my ID badge finally works, which means that I’ll be able to access most of the doors in the Center for Human genetics Building !!!
Finally Friday was more of an analyzing day. First, I scanned twelve plates unto the computer so that they can be run through on the 7900 machine. After a few minutes of waiting, I placed the plates in the cold room and analyzed the results to see if my PCR from the previous day was successful. To analyze the results, we used the program SDS 2.2.2. It generated graphs of the data, which is then submitted to a statistician for further evaluation. As it turned out, all of the plates that I completed were great. When I finished looking at the data that I produced, Chris showed me some useful tools in trying to find known mutations. There is a website called http://genome.ucsc.edu/ that is very helpful in doing so. We also used the Biomek robot to prepare for the PCR. Chris promised to show me how to use the PCR on Monday, so I’m very thrilled for that. Overall, this week was wonderful !!!!!
Week Two!
I have also met a few other people in the lab this week. There are a few graduate students, who have just gone to California for a big conference, and a few undergraduate students from a different summer research program at Duke. Some are in the MOB program (not sure what that stands for!), and one is a Duke senior working on a big project that will be presented in San Diego in November. There is also a Howard Hughes undergraduate, though I have not had the chance to meet her just yet. All around me there is interesting work going on, and I've been looking in during my many TBS rinses!
When I wasn't working on data analysis Dr. Devlin and Dr. Markert spent time teaching me about other important concepts in the lab. Patients with atypical DiGeorge are born with some T cells, but because the babies are athymic the T cells can't recognize self from non-self. These T cells are likely to bind with self antigens and proliferate. The result is large oligoclonal populations and a terrible rash for the patient. Spectratyping is a tool that uses RNA to show you the diversity of the T cell receptors. An atypical patient might show a large T cell population but with spectratyping you'd find they had only a few oligoclonal populations, as opposed to a normal very diverse population.
On Friday, David, the undergrad, brought his samples to the DNA sequencing lab so I tagged along. Instead of just dropping off the tubes with the lab, we got to sequence it ourselves which involved combining the DNA with a mix of primers, enzymes and nucleotides (some fluorescent) and loading the samples into the machine, the very very expensive machine. The actual process was a little anticlimactic but learning about the how the process and machinery all worked was interesting. All in all it was another good week. Soon I'll get to do more bench work along with the continuing data analysis, so I'm looking forward to that. Hope everyone is having a great weekend!
it happened
Cheers!
Friday, June 22, 2007
insert intriguing title here
Plant Nerd
I've been thinking - I've always considered myself an animal-person (my house = perpetually overcrowded zoo / animal hospital), and I am...but this summer, I realized how incredibly amazing/intruiging/unbelievably complex plants are! Since I was young, my parents bought flat after flat of plants from Lowe's and also rescued unwanted ones from various people moving back to Japan, and I had always wondered what it was about plants that fascinated my parents. I viewed plants as boring because no matter how hard I worked, they never seemed to interact with me - they don't play fetch, hop onto my lap, beg for carrots, or curl up in my hair. But I realized this summer that plants aren't just those green, crunchy, photosynthesizing "things" that we eat in salads - they are incredibly diverse and have so many different mechanisms for survival that have evolved throughout their history. I'm kind of disappointed about not realizing this earlier...It's kind of like when you're in the same class with a person for a whole year, but you never really get to know her until the last day when you realize how charismatic she truly is...and then you want to hit yourself for not taking the opportunity to get to know her better when you had had the chance to for the whole year. But I'm happy because I have the rest of my hopefully long life to get to know plants better.
I can't believe it's already been two weeks! I've now begun stopping to examine random plants while I'm running and looking at the distance between the anther and stamen (I'm really interested in that trait!), and I'm now performing experiments with the various begonias, Saint Paulias, and Torenias around my house to see if I can cross-polinate and/or self them. I feel like such a plant nerd...and I love it!
 Hi guys!
Hi guys!This week, I've been prepping for my project, which is the study of transfer rates of plasmids between bacteria in batch bioreactors. A bioreactor (that one is quite complicated-->) is a vessel where environmental conditions like the amounts of different gases, flowrates, temperature, and pH can all be controlled; but for our experiment, we are going to use bottles with vaccum seals instead. Next week, in the bioreactors, we are going to test six different concentrations of bacteria. We have already made triplicates of each combination of bacteria so that if one sample is faulty, then we still have two other ones to study, and we'll study how they're similar to or different from each other, of course. Right now, I have 34 bottles of solutions waiting to be inoculated with the bacteria on monday. I'll come in on sunday to watch my mentor prepare the bacteria so that I can learn how for later; something I've learned this week is that scientific research has no time constraints! Anyway, today, I also have to make a calibration curve comparing different amounts of toluene.
 The graph will become our standard, and the measurements we obtain from the bioreactors will be compared to the calibration curve. The optimal slope should be .999; hopefully we'll get that number today. My mentor Ruoting told me that I'll be very busy next week with the 34 bioreactors, so I'll be having an extremely productive few days in the lab! I'm really excited to see how the data turns out because we'll be testing to see whether an E-Coli conjugate with P. Putida's plasmid exists.
The graph will become our standard, and the measurements we obtain from the bioreactors will be compared to the calibration curve. The optimal slope should be .999; hopefully we'll get that number today. My mentor Ruoting told me that I'll be very busy next week with the 34 bioreactors, so I'll be having an extremely productive few days in the lab! I'm really excited to see how the data turns out because we'll be testing to see whether an E-Coli conjugate with P. Putida's plasmid exists.Have a lovely weekend,
Annie
PCR power

Yes, so I am still working on my project, which will involve 27 primer pairs and 12 sea urchin individuals. If you do the math, that's 324 different base pair sequences. Here's the process: PCR, gel electrophoresis, DNA purification, specs, and sequencing. For 324 different tubes. Needless to say, it will take some time, but I am almost finished PCRing all the tubes. Yesterday, I did an 8 by 12 plate of PCR reaction. Once we could get it to work, I used an automatic pipette to alloquate the primer master mixes into the 96 tubes. Surprisingly, PCR involves more math than I thought. It wasn't difficult, but I was actually forced to use a calculator. WHen I am creating the PCR master mixes, sometimes I feel like I'm cooking, putting together a recipe.. 4 ul of sugar, 25 ul of water, and 750 ul of baking soda. Delicious. But like cooking, you have to be careful about not putting in the correct amount of each ingredient. PCR is a reaction that involves a lot of different components, and depending on how many tubes you have, you master mix will be correspondingly bigger or smaller. The lab even has its own blender- the tabletop vortex that mixes up the ingredients to make everything homogenous. Of course, all this "biological cooking" hasn't changed my mind about kitchen cooking... I still avoid it at all costs, but I am learning that scientists have to be alert and focused to get specific reactions like PCR right. If you're not careful, you could spoil the batch and waste ingredients while you're at it. Overall, it's been an interesting week.
Thursday, June 21, 2007
In the Field- Ecology
I started the week off with a shopping trip to Sears, Lowes and many more hardware stores to buy products for Eileen's project. Then, I spent more time helping out Eileen (a grad student) with her chambers that collect gases given off by plants. Wednesday and Thursday I stayed at the field most of the day to help install the chambers. I also helped Dr. Wright collect data on mono culture (one species of a plant) plants and if they had more herbivory than many species of a plant. We are still working on that project. I had lots of fun so far this week and I hope everyone is having a blast as well.
Baninder
Tuesday, June 19, 2007
Nicole and the "Drunken Mouse"
 r about an hour, most times featuring two mice for convenience, and the analyst is supposed to focus on just decoding one mouse behavior at a time. When I began watching the mice, of course their behavior was "different", even strange, but as I approached the time to analyze that small brown mouse in arena 20, the adjectives strange and different became better suited for use in understatements.
r about an hour, most times featuring two mice for convenience, and the analyst is supposed to focus on just decoding one mouse behavior at a time. When I began watching the mice, of course their behavior was "different", even strange, but as I approached the time to analyze that small brown mouse in arena 20, the adjectives strange and different became better suited for use in understatements. Post-injection, the mouse would sit up on its hind legs and rock from foot to foot as if it was "tippy", which is the word designated to describe this behavior in decoding. After rocking and leaning his/her head against the glass walls of the arena surroundings for a few seconds, she would proceed to play it cool as her little mouse body would erratically fall into the center of the arena. She had a lack of balance in her walk, sniff, and stance that could best be imagined if compared to an ad on the effects of alcohol consumption at its worse....

Monday, June 18, 2007
Number Systems in Babies and Adults
The thing that I am most interested in scentifically right now is early childhood development. Anyone who knows me is aware that I love kids. I babysit regularly; it is not just a job but an important part of my life. I have worked in the preschool at Friends School for a community service credit, and learned how to deal with kids that have simple A.D.D. or even selective mutism. Last year I interned at Frank Porter Grahm both in the childcare center with the two and three year olds (again, learning about development while working with kids who had DiGeorges and high functioning autism) as well as on a research project, First School, which looks at ways to improve education in children ages three though eight.
Working in Dr. Brannon's lab fits right in with that line of interest. Today Sara, my mentor, told me about the two ways babies and adults process number. The first system is the analogue system, which is essentially the approximation system. If there are 20 peanuts on the table, you might approximate that there are about 15-25 peanuts. If there are 200 peanuts on the table, that margin of error goes up to say, 150-250 peanuts. It also describes if you hold two weights of different sizes in your hand. With a one to two ratio, you would have no problem saying which was heavier. With a 51 lb weight and a 52 lb weight you might have more trouble.
The object tracking system is the other system that we use. Most adults can keep track of about four things. Sara had me do a little test online (Here it is if you want to try-keep track of the dots that blink at the beginning-http://research.yale.edu/perception/oba/MOT.mov). Although four is the limit for most adults, people like air-traffic controllers can track up to nine or so objects on a screen. Infants, on the other hand, we think can only track three. Infants could distinguish one from three, or two from three, but not three from six. Somehow, that conversion between analogue and infant tracking failed. We are now doing studies that look more into this. Although it is somewhat confusing, as I learn more about it I understand it better.
As usual, the lemurs are quite adorable. I found out today that their names are themed according to species. The ring-tailed lemurs are Greek names (Aracus, Licinius, Teres), the mongoose lemurs Mexican (Miguel, Pedro, Rosalinda, Guillermo), the blue eyed lemurs after blue-eyed movie stars (Tarantino, Witherspoon, etc.). The list goes on. For those of you interested, the website: http://lemur.duke.edu/
Linda
A little about myself:
I am pretty much the most clumsy and uncoordinated person ever; I bump into things and bruise easily. Fortunately, this has not proved to be catastrophic in my lab yet (little do they know). I fear bugs and tsetse flies and tapeworms and generally all parasitic protozoa. Around new people, I tend to be reserved, polite, and friendly. With people I know well, I'm generally hyperactive-crazy, incoherent, and hilarious. I loved to pounce on my friends in fits of tigger-worthy joy from behind...up until I was side-stepped one day, and thereby fell to the ground on my bum with an audible thump and acquired the condition known as coccydynia (inflammation of the tailbone, self-diagonsed off medecinenet.com). I can find the most random things hilariously funny and I love to laugh and make other people laugh, even if its at me and my awkward antics or germophobicness or hypochondriacness. I love to get lost in a good book, and I am impatiently anticipating the arrival of the last Harry Potter book! I love to reminise about Arizona, sunsets, moutains, and stress-free days. I love swimming, because it's a no-contact sport and I love to break it down to club music when I think no one is watching. And as a final caution, I have a voracious appetite for everything that does not contain trans fat or carcinogenic grill marks, so hide your food.
About my lab!:
I am working in the Experimental Surgery department in the Medical Sciences Research Building with Dr. William (Bill) Parker as my mentor. The project I will be working on involves a mouse that has been living in a sterile bubble for the duration of two years. It has not been exposed to any pathogens or bacteria, except for a certain strain of e. coli bacteria that has been dripped into a spout extending from the bubble structure. After this period however, two different strains of e. coli were discovered: a fast-growing strain and a slow-growing strain. The hypothesis I will be testing is that when inside the small intestines of the mouse, these bacteria had been growing at the same rate. From a broader perspective, this experiment could possibly further tap into the pool of knowledge regarding bacteria behavior and evolution.
So far, I have been learning some laboratory techinques and procedures from Mary Lou so that I will be well prepared for the experiment. I have entered the realm of preparing mucin (mucus from a pig's intestines) and BSA (Bovine Serum Albumin Fraction), running dialysis, concocting media cocktails, incubating a certain mysterious "bacteria 315", and preparing stick well cell cultures to test the bacteria growth on BSA versus BSA+ Mucin. Hopefully everything will turn out right!
Ta ta for now!
Sunday, June 17, 2007
It has been a whirlwind first week. On Monday, the first day, instead of being picked up by the usual one escort to take me to the lab I'd be working in, four people arrived. They were all quite enthusiastic and got me excited about spending the summer working with them. Once I arrived at the lab, I was given a tour and an introduction to what they do and what I'd be studying for the next seven weeks. They research how polycyclic aromatic hydrocarbons (PAHs) affect the aryl hydrocarbon receptor (AHR) pathway in zebrafish and killifish.
By the second day, they had me working with a graduate student studying zebrafish hatchlings. These hatchlings had been dosed with PAHs and we looked at them under a microscope, noting the impact of the PAHs. We were especially focused on the zebrafishes' development of pericardial edemas from their exposure to PAHs.
Another day I got to help spawn killifish. To do this, you hold the females and rub their stomachs with your thumb to make their eggs come out onto your gloved hand. You add the eggs to a jar and then fertilize them with a male fish. The first time that I got to help with this though, we came across a surprise. The person I was working with, Cole, was spawning a female fish and got two already fertilized eggs among many regular ones. Everyone in the lab assured me that this never happens. Eggs are supposed to be fertilized only after they leave the mother. They looked at these two eggs under a microscope and figured that it had been about a week since they were fertilized. It's a mystery to everyone in the lab as to how this happened and I'm interested to see what we discover about them as tests are run as they get older. For now, they are living in the lab and have been nicknamed Jesus Fish 1 and Jesus Fish 2.
I'm very excited to be working in this lab. It's already proven to be a very interesting week.
Lots of PCRs
I also learned to use the spec machine on Friday. This allows you to look at how efficient the DNA purification after PCR was. It measures the concentration of DNA vs. protein, as well as the content of DNA vs. other junk that you don't want. You pipette a tiny droplet of the mixture onto a small metal surface and the machine uses light rays to read the DNA concentration in the water.
Overall, I had a very exciting week. I have learned so much, and gotten so much experience. It was a rush of different protocols and vocabulary, and I can't wait to learn more next week and the rest of this summer!
Wandering Old Rats
I play a lot of sports, and I'm usually in one for at least 2 or so seasons a school year. I've done Cross Country, Swimming, and Track, and I'm starting tennis in the summer/fall. And I am totally open to playing tennis with someone who knows how some weekend!
I also enjoy listening to and finding music, most of my favorite bands being of the British indie sort, though I really like almost everything and I enjoy dabbling in a variety of genres (even opera, minimally!) Oh and you should check this video out--it's this absolutely amazing opera performance by this mobile phone salesman from Wales. Most of you have probably already seen it, but if you haven't, it's worth seeing. http://youtube.com/watch?v=1k08yxu57NA
My lab this year deals in neuroscience. I work with Melissa Glenn, a postdoc in Dr. Christina Williams' lab. I'm coming in at the end stages of a 2-year study on the effects of prenatal choline exposure levels on neurogenesis, memory, and explorative tendencies in aging rats who have been exposed to different levels of choline during the prenatal stages. So far, I've done everything from observing rat maze tests to watching perfusions and brain extractions, to moving delicate slices of rat brains into appropriate containers. The research that this lab does really has the potential to be quite revolutionary. Eventually, it is plausible that research like this can help improve quality of life drastically for the aging population. These old rats (90-something in rat years) and their seemingly random wanderings have the capacity to change lives! Who would have thought?
It looks like I'm in for an exciting summer! I'm really happy that I am a part of this possibly groundbreaking study.







